Irving, CA, November 13 (RHC), -- Scientists are creating new technology to selectively and rapidly detect bacteria directly from a blood sample within 1.5 to 4 hours and are able to control infections and other major blood illnesses, according to a report published in Nature Communications Journal on November 13.
Blood stream infection or sepsis is a major health problem worldwide, with extremely high mortality, which is partly due to the inability to rapidly detect and identify bacteria in the early stages of infection.
The initial treatment with empirical broad-spectrum antibiotics not only is inadequate but also encourages antibiotic resistance. It is widely recognized that effective detection and monitoring of patients to diagnose a blood stream infection (BSI) at an early stage has a profound effect on survival rates. However, the present gold standard, bacterial blood cultures coupled with susceptibility testing for drug resistance, requires days to obtain a result.
Inevitably, the existing methods typically require expensive equipment and lengthy, complex sample processing (for example, cell lysis, nucleic acid extraction, centrifugation, magnetic separation, washing and signal amplification) for target purification and enrichment, which not only results in significant loss of rare target organisms, and therefore contributes to a high false-negative rate, but also limits their widespread use especially in a point-of-care setting (for example, in an ambulance).
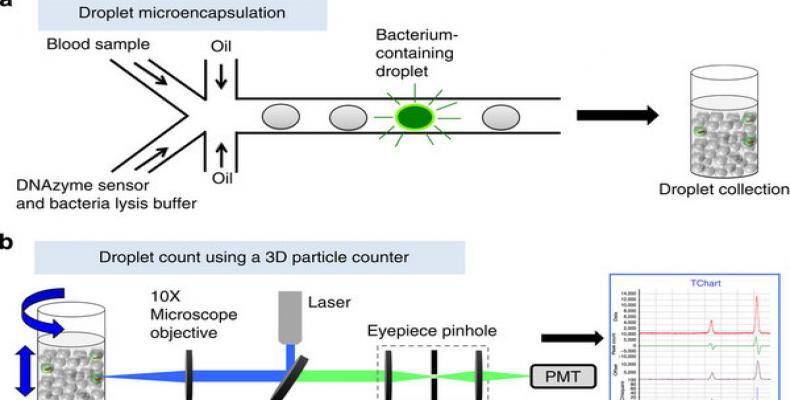
But now, researcher Dong-Ku Kang, M. Monsur Ali, Enrico Gratton and colleagues from the University of California -Irving, present a new technology termed ‘Integrated Comprehensive Droplet Digital Detection’ (IC 3D) that can selectively detect bacteria directly from milliliters of diluted blood at single-cell sensitivity in a one-step, culture- and amplification-free process within 1.5–4 h.
The IC 3D integrates real-time, DNAzyme-based sensors, droplet microencapsulation and a high-throughput 3D particle counter system. Using Escherichia coli (E. coli) as a target, scientists demonstrate that the IC 3D can provide absolute quantification of both stock and clinical isolates of E. coli in spiked blood within a broad range of extremely low concentration from 1 to 10,000 bacteria per ml with exceptional robustness and limit of detection in the single digit regime.
In pilot experiment, scientists tested their hypothesis that the confinement of bacteria in droplets enables single-cell sensitivity and reduced detection time.
“For detection of bacteria in blood, we needed to optimize droplet size: although smaller droplet sizes lead to higher target concentrations from single cells (which would increase the signal/background ratio and decrease the detection time), it is technically challenging to encapsulate blood contents including red and white blood cells into too small-sized droplets.” said a researcher.
Future integration of other sensing methods (for example, enzymatic assays, PCR and isothermal signal amplifications) with droplet microfluidics and a 3D particle counter may potentially serve as a platform for rapid detection and analysis of almost any type of low abundant markers in biological samples including cells (for example, bacteria, circulating tumor cells and stem cells), extracellular vesicles (for example, exosomes), viruses (for example, HIV) and molecular markers (for example, nucleic acids and proteins). In particular, the IC 3D system can potentially integrate fluorogenic substrates for beta-lactamases and carbapenemases, which would allow the detection of extended spectrum beta-lactamase-producing Enterobacteriaceae and carbapenem-resistant Enterobacteriaceae that are among the most prevalent antimicrobial-resistant pathogens.
This work is supported by the start-up fund from the Department of Pharmaceutical Sciences, Sue and Bill Gross Stem Cell Research Center and the Chao Family Comprehensive Cancer Center at UC Irvine. The project described was in part supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health.